CUDA-DTW
Subsequence Search under Euclidean Distance and Dynamic Time Warping

Algorithms
This supplementary website of our paper "CUDA-Accelerated Alignment of Subsequences in Streamed Time Series Data" provides additional material for the parallelization of Subsequence Euclidean Distance (ED) and Subsequence Dynamic Time Warping (DTW) on CUDA-enabled accelerators. The complete source code including the used datasets can be found on the left side at the corresponding github repository.
z-Normalized Subsequence Euclidean Distance
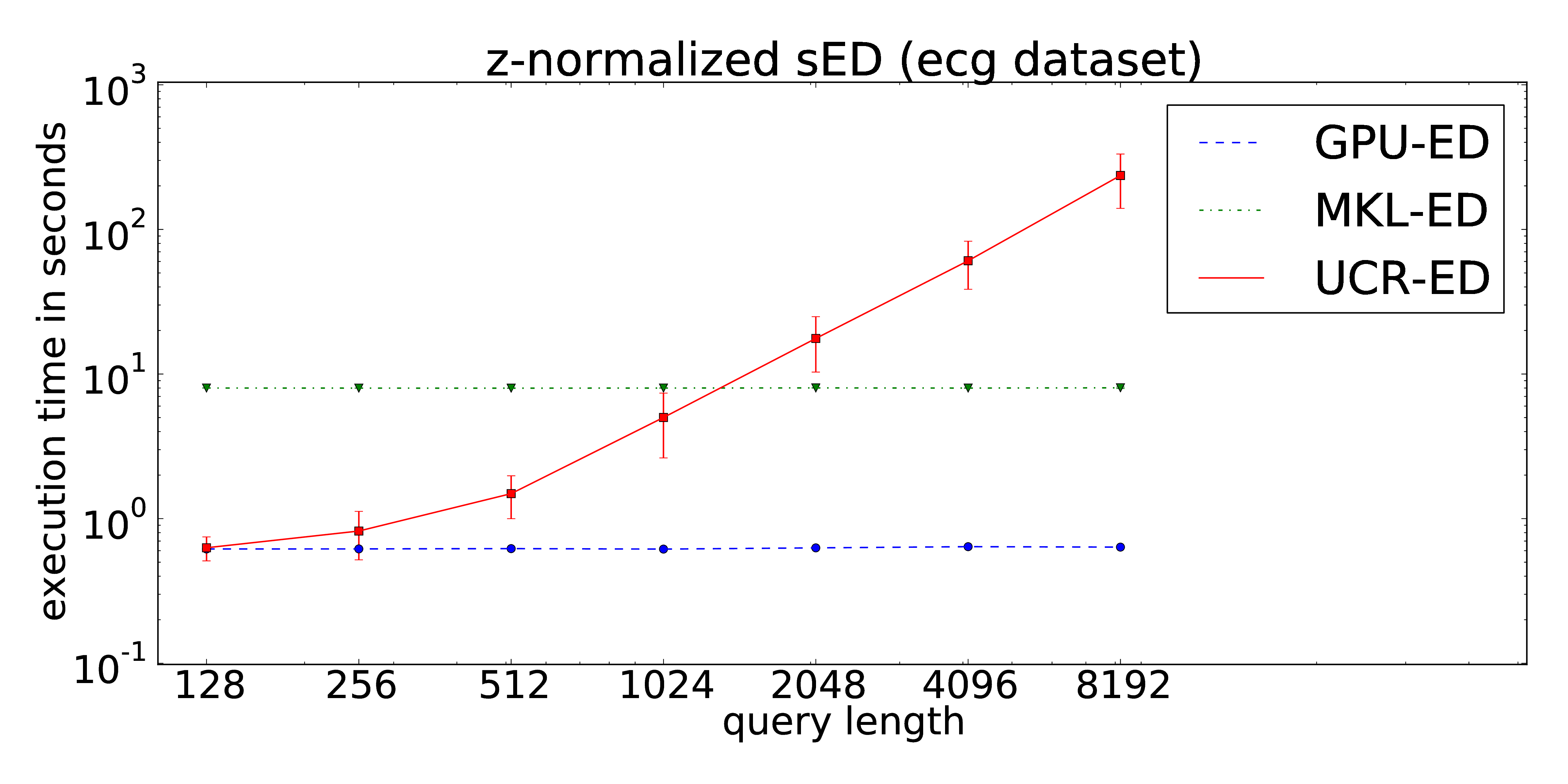
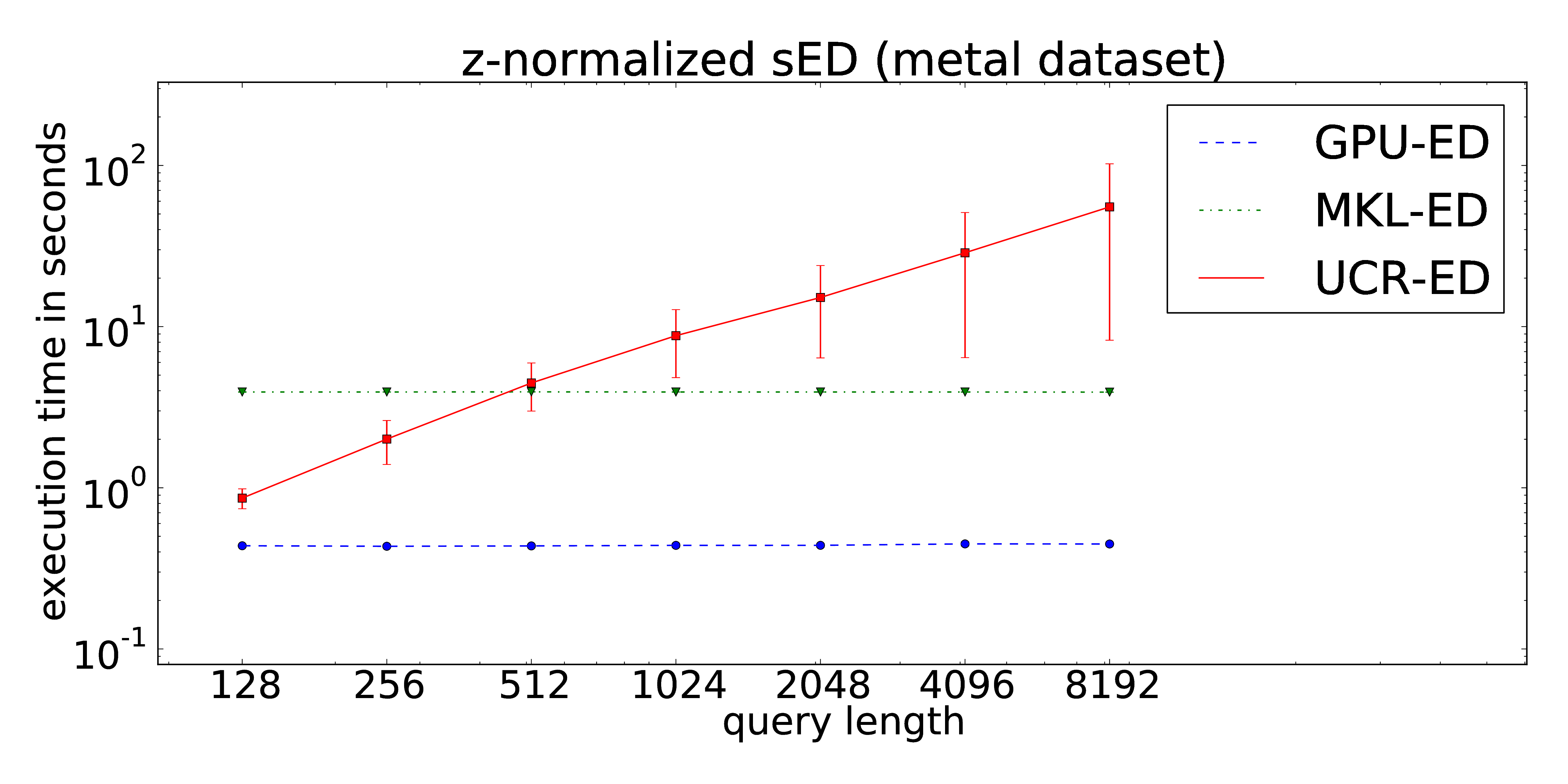
Our proposed implementation of the locally z-normalized alignment of time series subsequences in a stream of time series data makes excessive use of Fast Fourier Transforms on the GPU. The CUDA-parallelization features log-linear runtime in terms of the stream lengths and is almost independent of the query length. As a result, arbitrarily long queries can be performed without increasing of the runtime in contrast to the ED portion of the UCR-Suite.
 The core routines can be found at our github repository.
The core routines can be found at our github repository.
z-Normalized Subsequence Dynamic Time Warping with Sakoe-Chiba Constraint
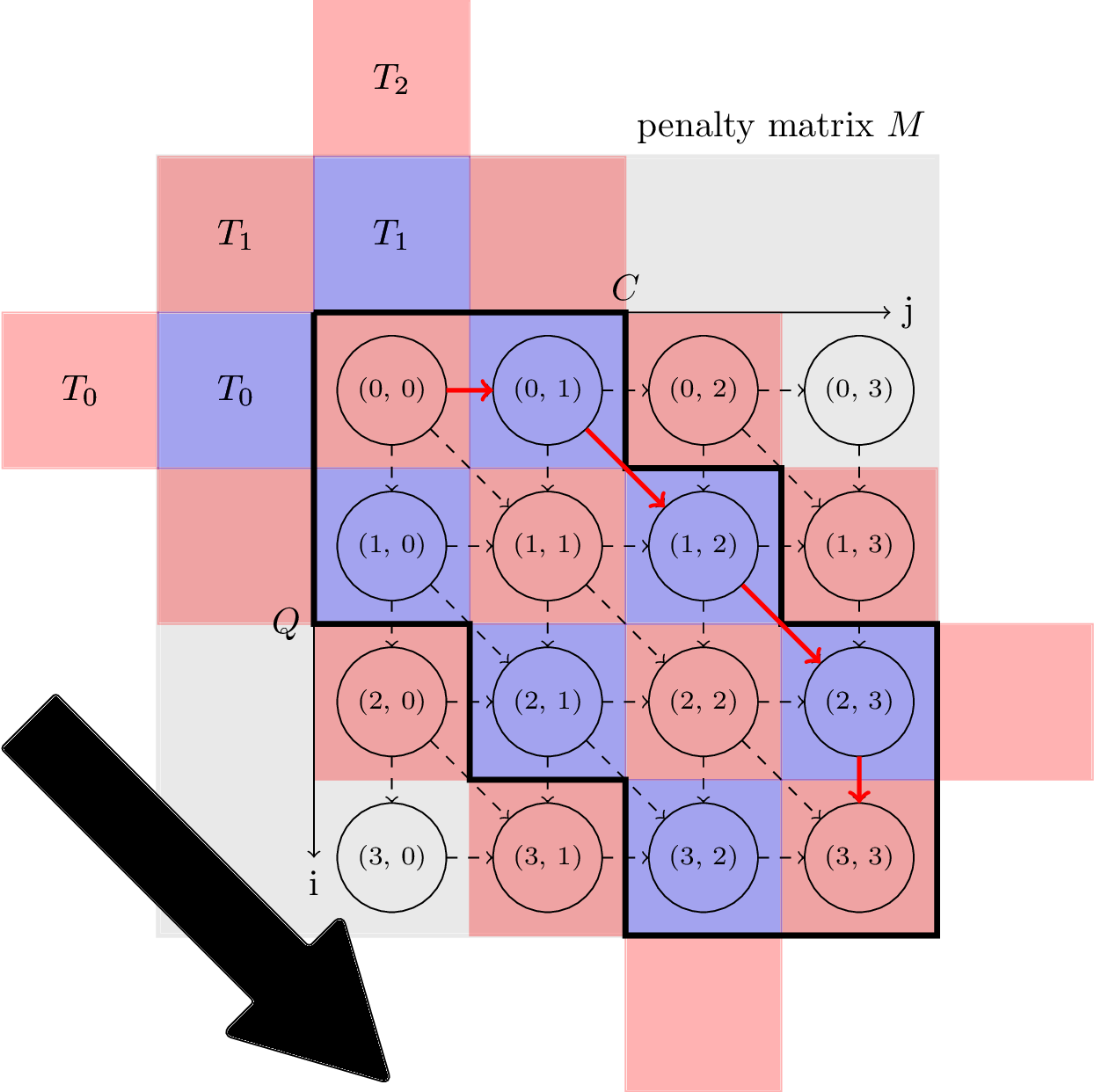 The proposed parallelization scheme of Constrained Dynamic Time Warping uses wavefront relaxation of the corresponding Sakoe-Chiba band. Furthermore, LB_Kim and LB_Keogh lower bound cascades are provided to prune unpromising candidates. Our CUDA version performs one to two orders-of-magnitude faster than the DTW portion of the UCR-Suite.
The proposed parallelization scheme of Constrained Dynamic Time Warping uses wavefront relaxation of the corresponding Sakoe-Chiba band. Furthermore, LB_Kim and LB_Keogh lower bound cascades are provided to prune unpromising candidates. Our CUDA version performs one to two orders-of-magnitude faster than the DTW portion of the UCR-Suite.
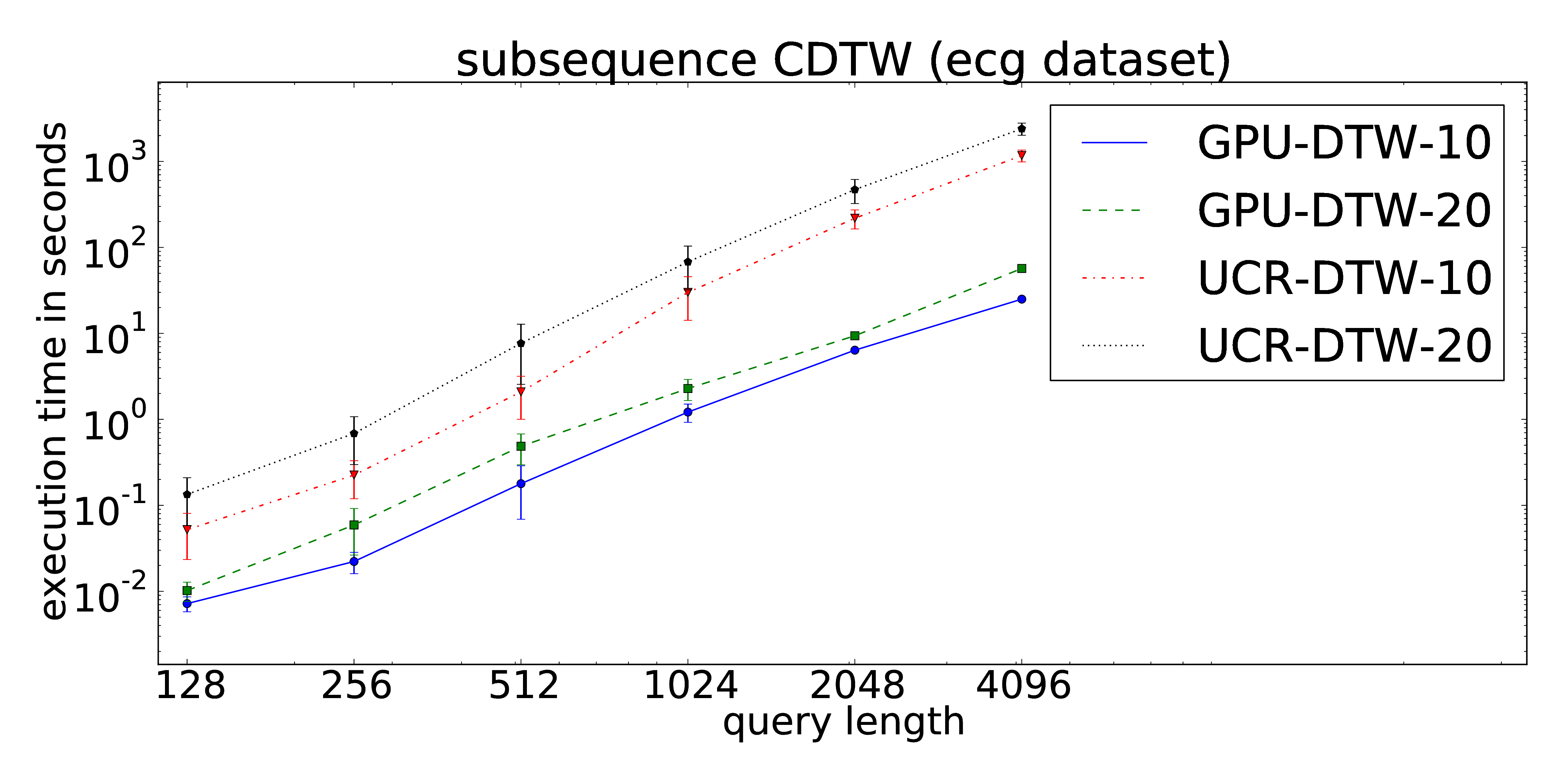
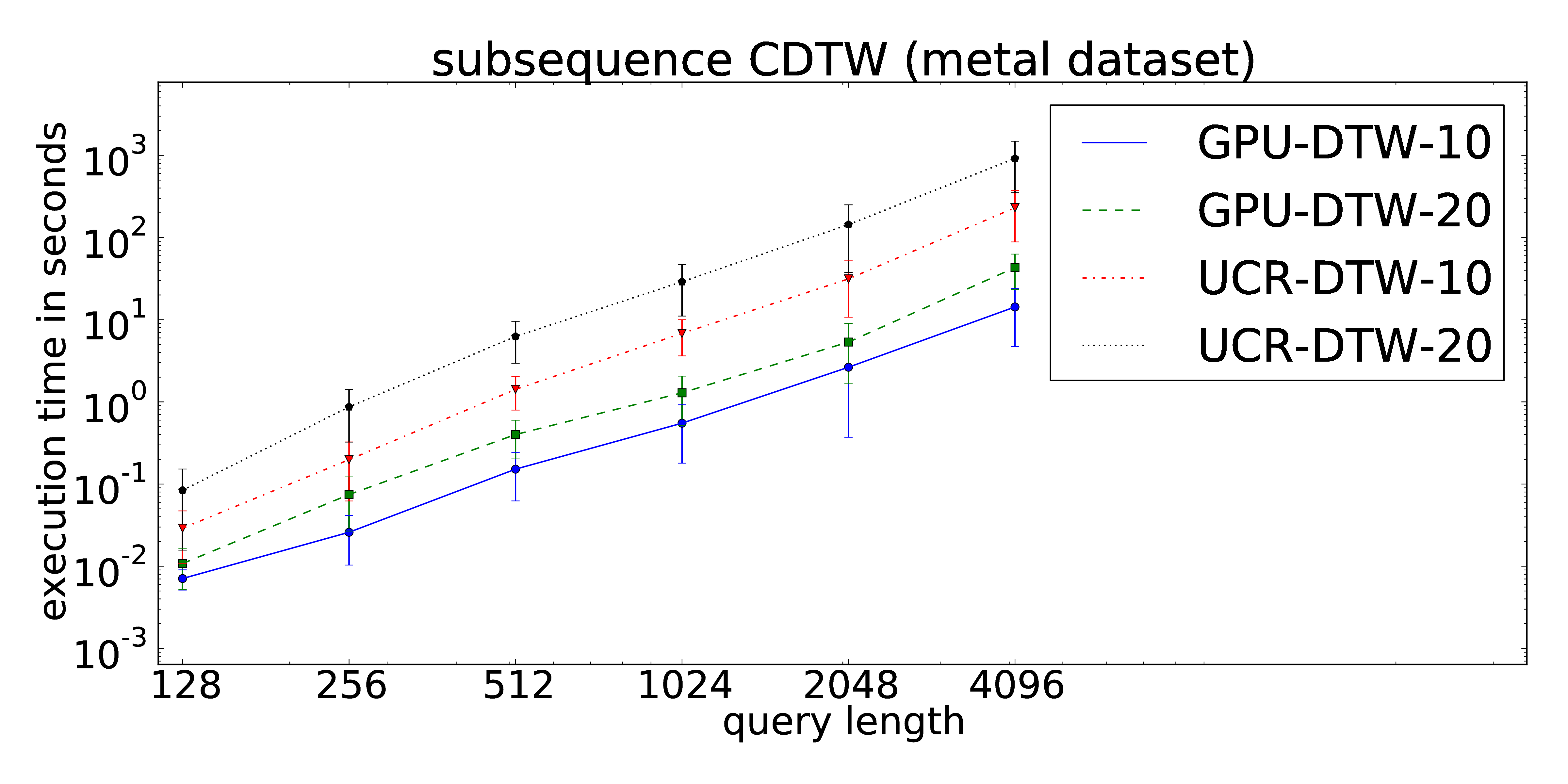 The core routines can be found at our github repository.
The core routines can be found at our github repository.
Datasets
ECG
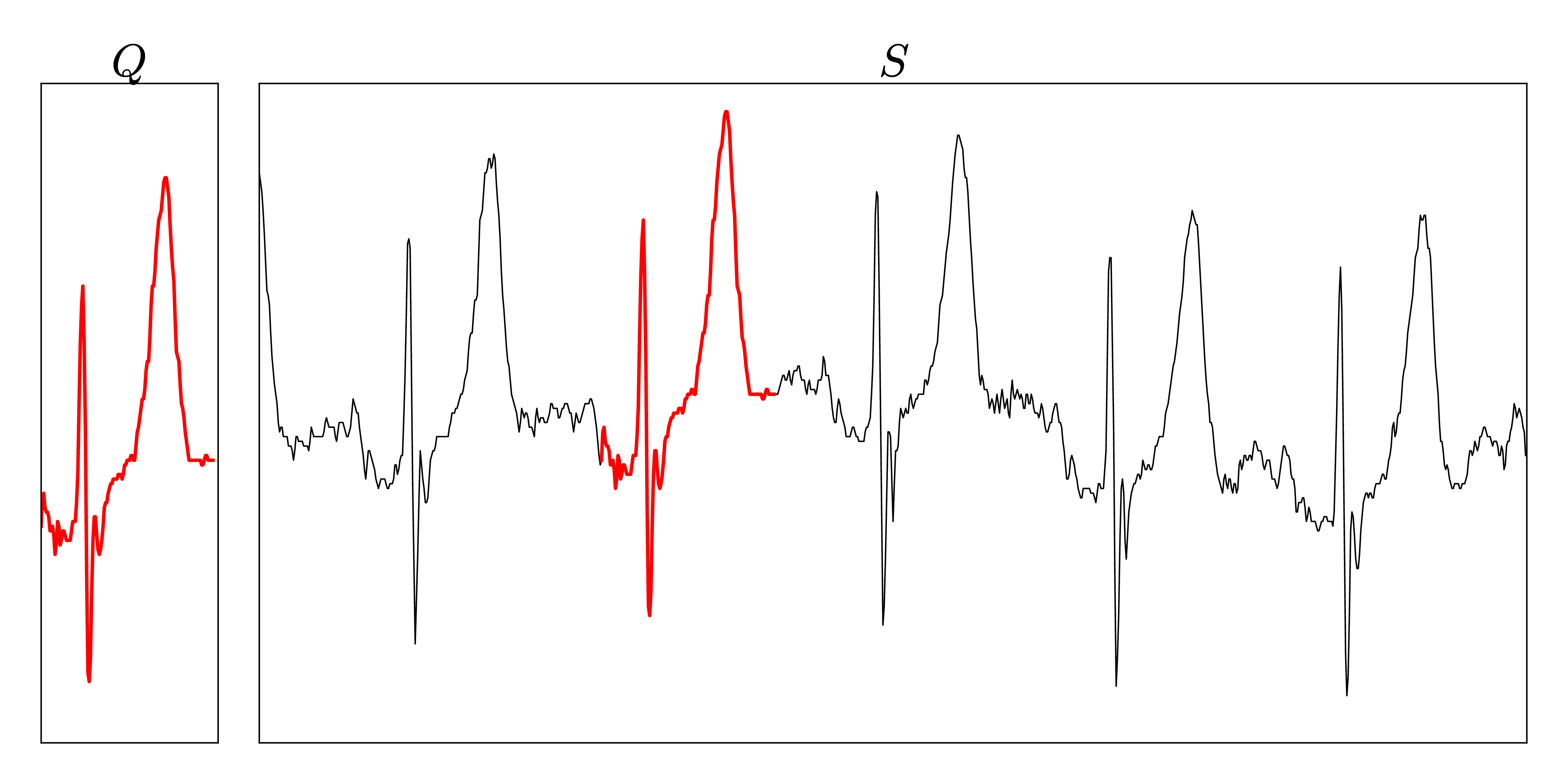
The dataset consists of approximately 22 hours streamed electrocardiograms (20,140,000 data points) and was used during the evaluation of the UCR-Suite by Rakthanmanon et al. The full dataset can be downloaded at the supplementary website of the paper. We reuse this dataset to ensure a fair comparison between the UCR-Suite and our implementations. The use of windowed z-normalization is advisable due to the time-dependent offset of the stream. The used queries and streams can be found here.
Metal
The dataset consists of 1,000 pairs of thickness signals measured during the hot rolling of metal bands in steel mills. The obtained time series are used for quality monitoring during the production process. Each time series is measured twice to avoid accidental mix-ups. Hence, we can access two individually recorded streams (both of about 11 Mio. data points) -- one for the extraction of the queries and one for the target data stream. The full dataset can be downloaded here. Analogous to the ECG signals, the data exhibits time-dependent offset shifts, too. Hence the use of windowed z-normalization is advisable.